The human genome contains a total of ~3 billion DNA base pairs. An estimated 1-2% of the genome codes for proteins, which influence everything from hair and eye color to disease susceptibility. The remaining base pairs are called “noncoding” and contain less well-understood instructions about when, where and how much protein-coding genes should be produced or expressed.
These instructions are critical to healthy development and lifespan, and mutations in noncoding DNA can affect physical traits like height and cause diseases such as cancer. Understanding how these noncoding sequences and their variants function could help explain if and when a person could be at risk for disease.
To unlock a deeper understanding of noncoding DNA, Principal Investigator David Kelley and scientist Vikram Agarwal started a collaboration with a team of DeepMind researchers led by Žiga Avsec. Together, they developed a new deep learning model named Enformer to better predict how DNA sequence determines gene expression.
New technology can “see” noncoding DNA further from the gene
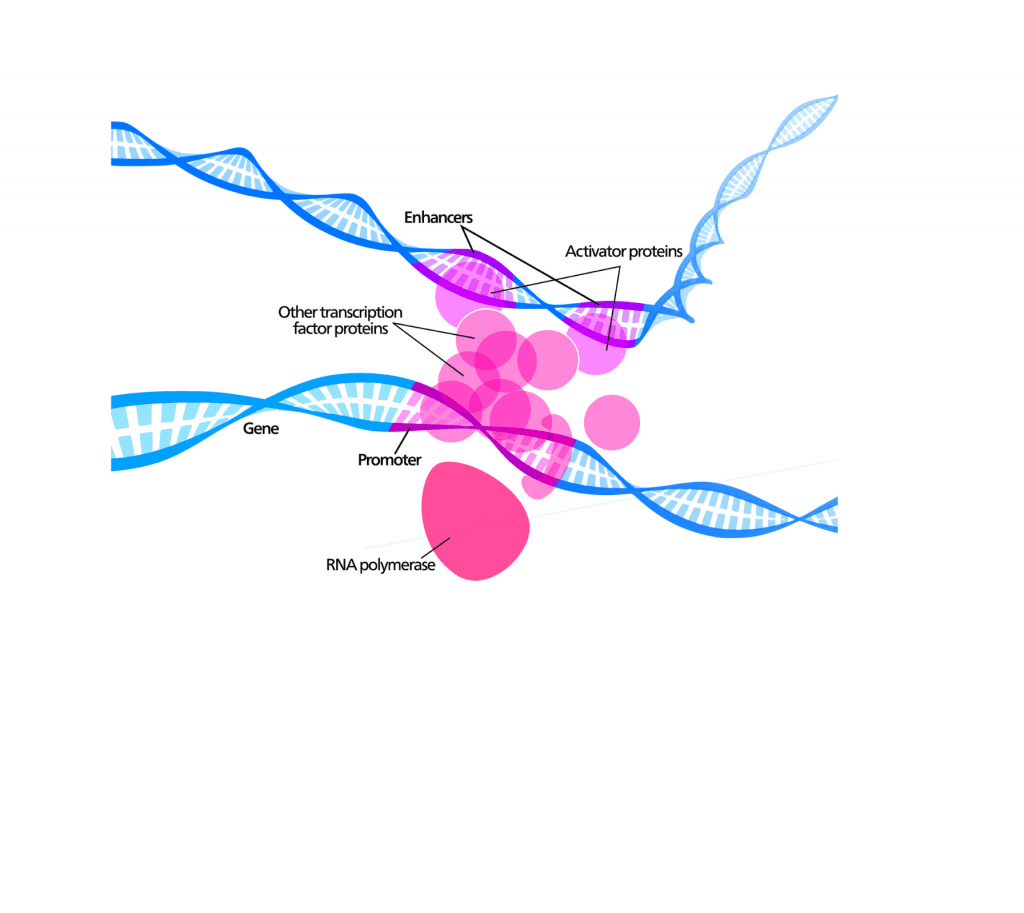
Enformer’s neural network architecture is based on Basenji, a previous program published by David’s group that can predict gene regulatory activity (one main function of noncoding DNA) from DNA sequence. Basenji considers ~40,000 local base pairs to make its predictions, but many known regulatory elements occur at distances greater than that. These include “enhancers,” which are noncoding DNA elements that can increase gene expression even at a million base pairs away from a gene.
The previous Basenji model could not see far enough from genes and missed many enhancer elements. So the team searched the machine learning literature for techniques that would increase this visibility and found a neural network layer called “transformer” that accomplished the goal. The name Enformer is a portmanteau of enhancer and transformer.
After demonstrating that the new models improve gene prediction accuracy from 0.64 to 0.71 Spearman correlation on gene sequences they’ve never seen before, the team studied the long-range behavior in depth, bringing in datasets generated by labs around the world to profile enhancer sequence activity to further validate the model. They went on to report more accurate predictions for how human genetic variants (or mutations) affect gene expression for both natural genetic variants in the population and designed mutations measured by reporter assays in the lab. The improvement represents a modest increase over previous methods, but it is well-supported by a diverse set of analyses.
An advance in genetic research
Enformer is a step forward toward making better use of noncoding DNA in genetic research. Since most human disease associations involve noncoding DNA, the advances the team made will help researchers determine the causal variants and the genes they affect in specific cell types. The team intends to collaborate with other researchers at Calico and beyond to ultimately help the scientific community improve their understanding of why one person might develop a disease while another doesn’t, and also offer insights into effective treatments.
“There is this untapped potential of disease associations that require new tools and strategies to understand,” said David. “Every association for which we can work out the details offers a seed that will hopefully grow into better understanding of diseases and how we might treat them to slow the aging process and improve quality of life.”
Read the paper published in Nature Methods and explore Calico’s other publications.
The image was created by Kelvinsong (own work) [CC BY 3.0], via Wikimedia Commons (modified). It illustrates the many diverse regulatory elements that influence gene expression, including enhancers that contact the promoter from a distance.